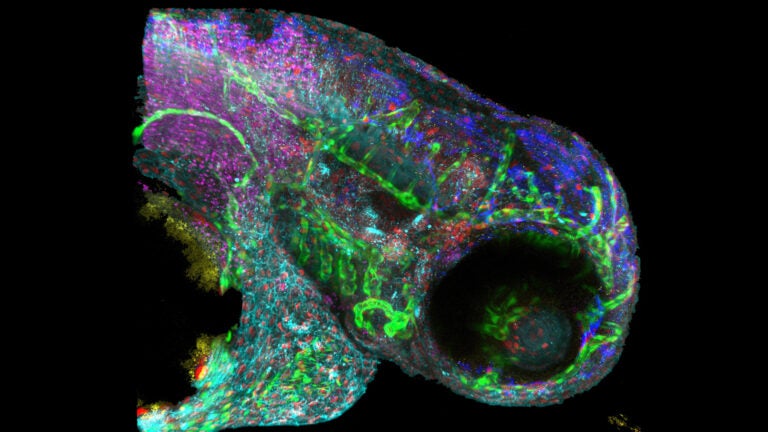
New technology improves complex imaging of biomolecules, such as this zebrafish embryo. (Photo/Courtesy of Francesco Cutrale)
New technology developed at USC could be used for diagnosing diseases
The technique known as Hyper-Spectral Phasor analysis makes finding biological molecules much faster and less expensive
https://www.youtube.com/watch?v=RvtrxhGRhlg&feature=youtu.be
A new image analysis technique developed at USC makes finding important biological molecules — including tell-tale signs of disease — and learning how they interact in living organisms much faster and far less expensive. Called Hyper-Spectral Phasor analysis, or HySP, it could even be useful for diagnosing and monitoring diseases using cellphone images.
Researchers use fluorescent imaging to locate proteins and other molecules in cells and tissues. It works by tagging the molecules with dyes that glow under certain kinds of light — the same principle behind so-called “black light” images.
Fluorescent imaging can help scientists understand which molecules are produced in large amounts in cancer or other diseases, information that may be useful in diagnosis or in identifying possible targets for therapeutic drugs.
Looking at just one or two molecules in cell or tissue samples is fairly straightforward. Unfortunately, it doesn’t provide a clear picture of how those molecules are behaving in the real world. For that, scientists need to expand their view.
“Biological research is moving toward complex systems that extend across multiple dimensions, the interaction of multiple elements over time,” said postdoctoral fellow Francesco Cutrale. He developed HySP with Scott Fraser, holder of the Elizabeth Garrett Chair in Convergent Bioscience and Provost Professor of Biological Science. The work was done at USC’s Translational Imaging Center, a joint venture of the USC Dornsife College of Letters, Arts and Sciences and the USC Viterbi School of Engineering.
“By looking at multiple targets or watching targets move over time, we can get a much better view of what’s actually happening within complex living systems,” Cutrale said.
A time-consuming process
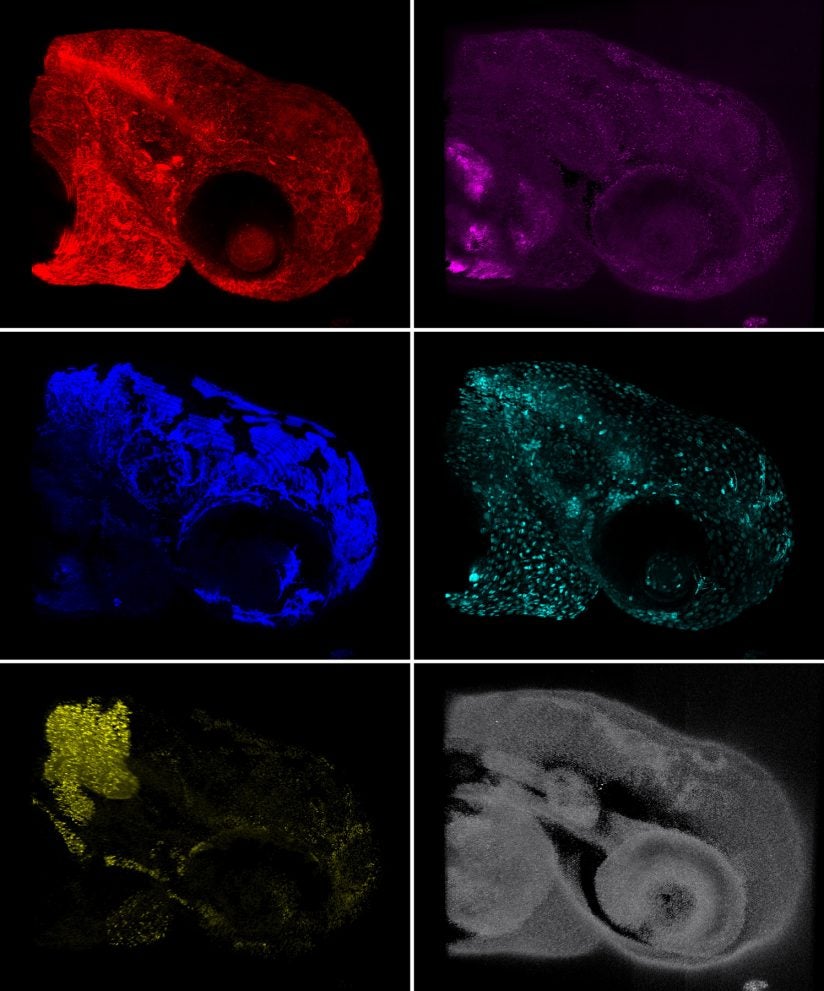
Currently, researchers must look at different labels separately, then apply complicated techniques to layer them together and figure out how they relate to one another, a time-consuming and expensive process, Cutrale said. HySP can look at many different molecules in one pass.
“Imagine looking at 18 targets,” Cutrale said. “We can do that all at once, rather than having to perform 18 separate experiments and try to combine them later.”
In addition, the algorithm effectively filters through interference to discern the true signal, even if that signal is extremely weak — very much like finding the proverbial needle in a haystack. Recent technology from NASA’s Jet Propulsion Laboratory can also do this, but the equipment and process are both extremely expensive and time-consuming.
“HySP uses much less computing time, and we don’t need the expensive imaging instrumentation,” said Fraser, who holds joint appointments at USC Viterbi and the Keck School of Medicine of USC.
In research published Jan. 9 online by the scientific journal Nature Methods, Cutrale and Fraser, along with researchers from Keck School of Medicine, Caltech and the University of Cambridge in the United Kingdom, have used zebra fish to test and develop HySP. In this common laboratory model, the system works extremely well. But what about in people?
“In experimental models, we can use genetic manipulation to label molecules, but we can’t do that with people,” Fraser said. “In people, we have to use the intrinsic signals of those molecules.”
Those inherent signals, the natural fluorescence from biomolecules, normally gets in the way of imaging, Fraser said. However, using this new computer algorithm that can effectively find weak signals in a cluttered background, the team can pinpoint their targets in the body.
Testing the process
The scientists hope to test the process in the next couple of years with the help of soldiers whose lungs have been damaged by chemicals and irritants they may have encountered in combat.
The researchers will extend a light-emitting probe down into the soldiers’ lungs while the probe records images of the fluorescence in the surrounding tissues. They will then use HySP to create what amounts to a fluorescent map and compare it with that of healthy lung tissue to see if they can discern the damage. If so, they hope to further develop the technology so it may one day help these soldiers and other lung patients receive more targeted treatment.
It might also be possible one day for clinicians to use HySP to analyze cellphone pictures of skin lesions to determine if they are at risk of being cancerous, according to Fraser and Cutrale.
“We could determine if the lesions have changed color or shape over time,” Cutrale said. Clinicians could then examine the patient further to be certain of a diagnosis and respond appropriately.
Cutrale and Fraser see the technology as a giant leap forward for both research and medicine.
“Both scientists at the bench and scientists at the clinic will be able to perform their work faster and with greater confidence in the results,” Cutrale said. “Better, faster, cheaper. That’s the payoff here.”
Additional scientists on the study include Vikas Trivedi of USC’s Translational Imaging Center, Caltech and the University of Cambridge and USC’s Le A. Trinh, Chi-Li Chiu, John M. Choi and Marcela S. Artiga.
The research was supported by grants from the Moore Foundation and the National Institutes of Health (R01 HD075605, R01 OD019037).
