
AI Sheds New Light on the ‘Code of Life’
While ChatGPT, Bard and other artificial intelligence tools keep writers, teachers and fans of the Terminatormovie franchise up at night worrying about various apocalyptic scenarios, another use of AI offers more hopeful outcomes.
Researchers at the USC Dornsife College of Letters, Arts and Sciences are using AI and other computational methods to redefine how scientists view DNA and give a clearer, more all-encompassing perspective on the “code of life.”
The knowledge revealed promises to transform scientific fields ranging from cancer research to drug design to sustainability.
Revealing a deeper complexity
In simplest terms, the genetic code is composed of four letters — A, C, G and T. The letters represent the nucleotides adenine, cytosine, guanine and thymine, which are part of the DNA double helix. These four nucleotide letters spell out the genetic code for all living things.
While this simple version of the code has done a serviceable job for decades, it doesn’t begin to fully reveal the complexity of DNA.
“We wanted to find a new way of encoding DNA that goes beyond the linear letter code,” said Remo Rohs, chair of the Department of Quantitative and Computational Biology at USC Dornsife. He and his colleagues published significant research, which used large-scale experimental data, earlier this year in the Proceedings of the National Academy of Sciences. They also published similar experimental data for a family of cancer-related proteins called forkhead box transcription factors in Nucleic Acids Research last week.
These and other research advances place the department among those at the forefront of the new USC Frontiers of Computing initiative, which aims to spur research and innovation in advanced computing technologies such as AI and machine learning, data science, blockchain and quantum information.
Researchers see DNA as more than a simple code
Rohs, professor of quantitative and computational biology, chemistry, physics and astronomy and computer science, and his team are looking to develop a more realistic and wholistic definition of the genetic code that includes “all structural variations and chemical modifications that we know of now or that could be discovered in the future,” he said.
These chemical modifications and structural variations that Rohs mentions range from small changes to the four nucleotides all the way up to major alterations that affect how DNA coils around itself and other molecules such as proteins.
These changes can affect which genes are active and which are dormant by allowing or blocking proteins from interacting with the DNA or reading the code.
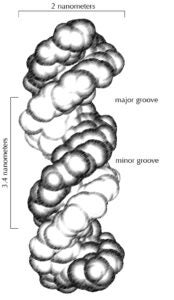
Rohs’ approach replaces the simple four-letter sequence with one that includes physicochemical groups in the major and minor grooves in the DNA double-helix.
So, what does that mean?
The DNA double-helix forms a twisted ladder shape. By virtue of the ladder’s twist, it has a wider, major groove and a narrower, minor groove. Depending on their size and shape, cellular molecules may have an easier time interacting with the DNA through one groove versus the other.
“Physicochemical” refers to both physical and chemical properties. Rohs’ method takes into account the various bumps and protrusions of the nucleotides and other DNA components and their physical accessibility within the two grooves. It also incorporates how the DNA components might react chemically with proteins. Taken together, this gives a clearer picture of how the cell’s machinery interacts with and interprets the genetic code.
For instance, a protein might normally bind to a section of DNA coded as AGTCATGGA, but if that section is tucked away in the minor groove, the protein might not be able to get close enough to bind. Or, if the protein and coded section have a strong chemical attraction, even if the coded section is tucked in tight, the protein might still be able to interact, but to a lesser degree.
Greater insights on DNA using AI
This is where Rohs’ team introduces AI, which learns a DNA-binding protein’s preference for certain chemical groups at specific physical locations in each groove of the DNA.
By accounting for these nuances, Rohs and his team reveal a more complete picture of what happens with DNA in living cells, one that extends far beyond the simple, linear, four-letter code. This, says Soheil Shams, chief information officer emeritus of biotechnology company Bionano Genomics Inc., is key to advancing computational research on the genome.
“One of the most important, yet challenging, steps in many computational biology applications, like machine learning, is how to represent biochemical information so it can be computationally processed,” said Shams, who graduated from the USC Viterbi School of Engineering with a master’s degree in 1986 and a PhD in 1992. “The proposed approach by Dr. Rohs and colleagues is offering a much more complete representation of the DNA sequence that should enable similarly more complete discoveries in interpretation of genetic variants as well as cancer research.”
Rohs’ method would help scientists understand why some genes are only partially active under certain conditions, or why the activity of some genes increases or decreases with age.
And this, Rohs says, opens doors for a range of beneficial research avenues.
“Using AI methods on a genome with chemical modifications and structural modifications will allow its applications in cancer and aging research, agricultural research, synthetic biology, chemical engineering, and drug design,” he said. “For example, certain types of cancers involve chemical modifications of DNA, aging correlates with the level of DNA methylation, and plant genomes undergo extensive chemical modifications compared to the genomes of other organisms.”
For their next steps, Rohs says, the researchers want to apply their work to DNA-binding proteins that control gene activity and predict how altering nucleotides — or substituting new, synthetic nucleotides — affects those proteins’ function.
“We want to predict binding preferences of gene regulatory proteins, called transcription factors, to DNA with chemically modified nucleotides and synthetic base pairs to improve binding characteristics and develop drugs that improve human health and disease,” he said.
Rohs’ work on the cutting edge of computational biology using AI could hold benefits for humanity that bots like ChatGPT could only dream of.
About the studies
The paper published in Proceedings of the National Academy of Sciences was led by Postdoctoral Associate Tsu-Pei Chiu and co-authored by former graduate student Satyanarayan Rao.
The paper published in Nucleic Acids Research was led by postdoctoral associate Brendon Cooper and co-authored by former postdoctoral associate Ana Carolina Dantas Machado, lab technician Yan Gan, and professor of biological sciences Oscar Aparicio.
The experimental project was part of the Michelson Center for Convergent Bioscience. Both studies were supported by the National Institutes of Health and the Human Frontier Science Program.
More about research undertaken in Rohs’ lab is available at www.rohslab.org.