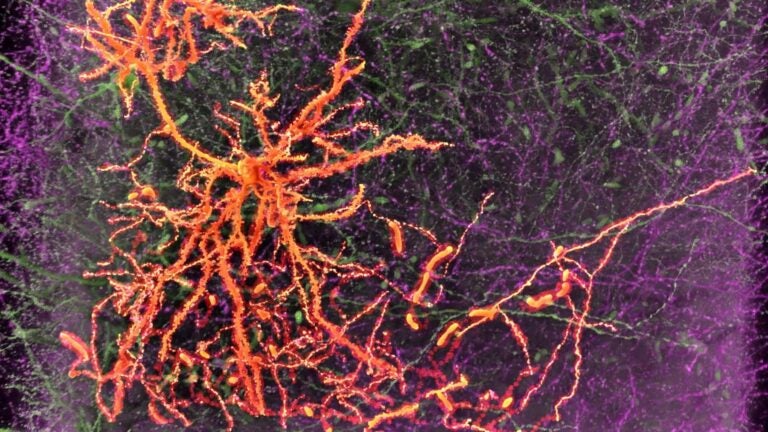
A neuron in the medial prefrontal cortex projects to the amygdala. (Image/Courtesy Hong-Wei Dong)
Brain cell study at USC gets $19 million National Institutes of Health grant
Researchers will identify and catalog each cell type in finer detail than ever before
The USC Mark and Mary Stevens Neuroimaging and Informatics Institute at the Keck School of Medicine of USC has received $19 million to help create a classification system for cells in the brain. Led by Hong-Wei Dong, associate professor of neurology, the team will join the BRAIN Initiative Cell Census Network consortium, a five-year multisite collaboration funded by the BRAIN Initiative.
Launched in 2013 by former President Barack Obama, the BRAIN Initiative is a public-private partnership led by the National Institutes of Health that unites federal agencies with foundations, universities and industry groups to develop and apply innovative technologies to revolutionize the study of the human brain.
In the present project involving mice, researchers will examine the organ in finer detail than ever before — combining anatomical, physiological and genetic approaches to paint a comprehensive picture of different cell types. It’s an extensive classification and mapping effort comparable in scale to the human genome project.
“When scientists first began studying the brain, we divided it into lobes,” Dong said. “Then we began classifying it into progressively smaller regions. Now we’re looking at groups of cell types and even individual cells.”
Joining the BRAIN Initiative, one of NIH’s most prestigious programs, means that Keck School of Medicine researchers will have the chance to impact diverse research activities and serve as a catalyst to other projects across the field of biomedicine.
We’re developing datasets and tools essential to illuminating the structure and function of the brain.
Arthur Toga
“We’re creating a classification system for a finer level of detail than we’ve ever tackled before,” said Arthur Toga, director of the institute. “In the process, we’re developing datasets and tools essential to illuminating the structure and function of the brain.”
Looking at all angles
The consortium’s overarching goal is to identify and catalog each cell type in the mouse brain, which requires tackling the problem from all angles: from connectivity mapping to observing molecular signatures, electrophysiological properties and functional relevance.
Dong’s team, already well established in the field through its revolutionary Mouse Connectome Project, will serve as the anatomy hub for the initiative. The group’s primary focus is on documenting and describing the structure and connections of various brain cells.
“We proposed to identify these cell types based on how they connect with other cells across the brain,” explained Houri Hintiryan, assistant professor of research neurology and a member of Dong’s working group. “A cell type will be determined by all of its connectional outputs and inputs.”
Dong is the principal investigator for the neuroanatomy analysis, based at the institute, and also provides support for two other projects: a connectivity and transcriptome analysis based at the Cold Spring Harbor Laboratory in New York and an epigenomics project based at the Salk Institute for Biological Studies in La Jolla.
For each project, Dong’s team will generate and analyze data, and provide informatics support. The group will employ a combination of the most advanced imaging technologies available, including cutting-edge microscopy and viral tracing methods.
Mapping the mammalian brain
This fall, Dong launched the USC Center for Integrative Connectomics, which houses a cross-disciplinary group working to develop a detailed connectivity map of the mammalian brain. Along with studying each cell type’s basic function, the center will explore larger cellular networks and disorders caused by connectivity issues. Answering these questions can reveal both how the brain functions when healthy and what factors drive disease.
“With all the advanced tools we now have available for use in animal models, we can selectively manipulate specific cell types to better understand their functional roles,” Hintiryan said. “Once we know how these neurons interrelate, it will facilitate our understanding of what goes wrong in various diseases.
“The hope is that we will have translational power for treating human neurological disorders,” she said.
The research is supported by the National Institutes of Health (U01MH114829, U19MH114821 and U19MH114831).
